Statistical Analysis
We first review some basic function of statistics: central tendency and dispersion before going into regression analysis.
Learning objectives:
summary statics
generate random numbers
estimate OLS
estimate time series models (ARIMA using arima, GARCH using rugarch, VAR using vars)
estimate panel data (plm package)
Simulation on random walk model and Roll model
Basic Statistics
Central Tendency
x <- 1:40
mean(x) ## [1] 20.5median(x)## [1] 20.5summary(x) #Five-number summary## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.00 10.75 20.50 20.50 30.25 40.00Dispersion
range(x) #Get minimum and maximum## [1] 1 40IQR(x) #Inter-quartile range## [1] 19.5var(x) #variance## [1] 136.6667sd(x) #Standard deviation## [1] 11.69045Random number
To generate random integers, we use the function sample().
We first generate 8 random integers between 2 and 10 without replacement:
sample(2:10,8, replace=F) ## [1] 8 3 5 10 4 7 2 9The next one generate 7 integers between 2 and 10 with replacement:
sample(2:10,7, replace=T) ## [1] 3 4 10 9 4 2 3In general, the sample() function can also be applied to extract random element from a vector.
namelist<-c("Mary", "Peter", "John", "Susan")
sample(namelist,2) # sample from this list## [1] "Mary" "John"To generate random decimal numbers, we use runif to get random uniform distribution. The following generates 2 random decimal numbers between 4.6 and 7.8:
runif(2, 4.6, 7.8) ## [1] 7.567968 6.025025To generate normal distribution, we use rnorm(). The following generate 4 random number that follows the normal distribution with mean being 0 and standard deviation being 1.
x <- rnorm(4, mean=0, sd=1) # standard nromal
x## [1] -0.91180710 0.62295902 0.92588317 -0.05877676Regression
We will be using the following data to show the regression. Note that we need to use dataframe.
x <- c(1,2,3,5,6,7,10,12,13)
y <- c(1,4,5,6,7,8,9,10,15)
z <- c(2,3,7,8,9,12,8,7,6)
df <-data.frame(x=x,y=y,z=z)Simple linear regression
The following code shows simple linear regression. The syntax is lm(y~x, dataframe).
SLR <-lm(y~x,df)
summary(SLR)##
## Call:
## lm(formula = y ~ x, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.9660 -1.2234 0.2618 0.7470 2.1627
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.5104 0.8756 1.725 0.128193
## x 0.8713 0.1134 7.686 0.000118 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.389 on 7 degrees of freedom
## Multiple R-squared: 0.8941, Adjusted R-squared: 0.8789
## F-statistic: 59.08 on 1 and 7 DF, p-value: 0.0001175Muliple linear regression
The following code shows multiple regression. The syntax is lm(y~x+z, dataframe).
MLR <-lm(y~x + z,df)
summary(MLR)##
## Call:
## lm(formula = y ~ x + z, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.8920 -1.2104 0.1329 0.8179 2.3030
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.25034 1.34526 0.929 0.388522
## x 0.85666 0.13321 6.431 0.000668 ***
## z 0.05167 0.19124 0.270 0.796060
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.492 on 6 degrees of freedom
## Multiple R-squared: 0.8953, Adjusted R-squared: 0.8605
## F-statistic: 25.67 on 2 and 6 DF, p-value: 0.001146Interaction terms
The following code shows multiple regression with interaction term. The syntax is lm(y~x+z+x:z, dataframe).
MLRDummy <- lm(y ~ x + z + x:z, df)
summary(MLRDummy)##
## Call:
## lm(formula = y ~ x + z + x:z, data = df)
##
## Residuals:
## 1 2 3 4 5 6 7 8 9
## -0.3080 0.9573 -0.3143 -0.7246 -0.1773 1.1230 -0.5891 -1.6242 1.6572
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.33212 1.96748 -0.677 0.5284
## x 1.59845 0.46581 3.432 0.0186 *
## z 0.64902 0.40027 1.621 0.1658
## x:z -0.12819 0.07789 -1.646 0.1607
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.316 on 5 degrees of freedom
## Multiple R-squared: 0.9321, Adjusted R-squared: 0.8914
## F-statistic: 22.89 on 3 and 5 DF, p-value: 0.002386Robust standard error
To have robust variance-covariance matrix, we have to use package sandwich to get the appropriate error term. Then we do the estimate through the package lmtest
First we install and load the packages.
install.packages(c("sandwich","lmtest"))
library(sandwich)
library(lmtest)Then we need to create the variance and covariance matrix: using the vcovHC().
To match the stata robust command result, we use HC1(). Then we perform coeftest based on specified variance-covariance matrix.
MLR <- lm(y ~ x + z, df)
coeftest(MLR, vcov = vcovHC(MLR, "HC1")) ##
## t test of coefficients:
##
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.250343 1.117886 1.1185 0.306129
## x 0.856663 0.194468 4.4052 0.004543 **
## z 0.051674 0.167447 0.3086 0.768062
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Time Series Analysis
Time Series data
Function ts convert any vector into time series data.
price <- c(2,3,7,8,9,12,8,7)
price <- ts(price)
price## Time Series:
## Start = 1
## End = 8
## Frequency = 1
## [1] 2 3 7 8 9 12 8 7We first define a time series (ts) object for the estimation. Note that ts is similar to xts but without date and time.
df <- data.frame(a = c(3,4,5),
b = c(1,2,4),
d = c(1,2,3))
df <- ts(df)
df## Time Series:
## Start = 1
## End = 3
## Frequency = 1
## a b d
## 1 3 1 1
## 2 4 2 2
## 3 5 4 3Extensible Time Series Data
To handle high frequency data (with minute and second), we need the package xts. The package allows you to define Extendible Time Series (xts) object.
The following code installs and loads the xts package.
install.packages("xts")
library(xts) Consider the following data for our extendible time series
date <- c("2007-01-03","2007-01-04",
"2007-01-05","2007-01-06",
"2007-01-07","2007-01-08",
"2007-01-09","2007-01-10")
time <- c("08:00:01","09:00:01",
"11:00:01","13:00:01",
"14:00:01","15:00:01",
"15:10:01","15:20:03")
price <- c(2,3,7,8,9,12,8,7)Now we are ready to define xts object. The code as.POSIXct() convert strings into date format with minutes and seconds.
df <-data.frame(date=date, time=time, price=price)
df$datetime <-paste(df$date, df$time)
df$datetime <-as.POSIXct(df$datetime,
format="%Y-%m-%d %H:%M:%S")
df <- xts(df,order.by=df$datetime) For conversion with dates only, we use POSIXlt() instead of POSIXct().
df <-data.frame(date=date, price=price)
df$date <- as.POSIXct(df$date,format="%Y-%m-%d")
df$price <-as.numeric(df$price)
price <-xts(df[,-1],order.by=df$date) ARMA
Autoregressive moving average model can be estimated using the arima() function.
The following code estimates an AR(1) model:
price <- c(2,3,7,8,9,12,8,7)
AR1 <- arima(price,c(1,0,0))
AR1##
## Call:
## arima(x = price, order = c(1, 0, 0))
##
## Coefficients:
## ar1 intercept
## 0.6661 6.1682
## s.e. 0.2680 2.1059
##
## sigma^2 estimated as 5.33: log likelihood = -18.34, aic = 42.68The following code estimates an AR(2) model:
AR2 <- arima(price,c(2,0,0))
AR2##
## Call:
## arima(x = price, order = c(2, 0, 0))
##
## Coefficients:
## ar1 ar2 intercept
## 0.8301 -0.3026 6.5911
## s.e. 0.3357 0.3638 1.7241
##
## sigma^2 estimated as 4.831: log likelihood = -18.01, aic = 44.02The following code estimates an MA(1) model:
MA1 <- arima(price,c(0,0,1))
MA1##
## Call:
## arima(x = price, order = c(0, 0, 1))
##
## Coefficients:
## ma1 intercept
## 0.7945 6.8862
## s.e. 0.4393 1.3412
##
## sigma^2 estimated as 4.936: log likelihood = -18.23, aic = 42.46The following code estimates an MA(2) model:
MA2 <- arima(price,c(0,0,2))
MA2##
## Call:
## arima(x = price, order = c(0, 0, 2))
##
## Coefficients:
## ma1 ma2 intercept
## 0.8292 0.1459 6.8262
## s.e. 0.3915 0.3245 1.4677
##
## sigma^2 estimated as 4.945: log likelihood = -18.14, aic = 44.28The following code estimates an ARMA(1,1) model:
ARMA11 <- arima(price,c(1,0,1))
ARMA11##
## Call:
## arima(x = price, order = c(1, 0, 1))
##
## Coefficients:
## ar1 ma1 intercept
## 0.3551 0.4889 6.6417
## s.e. 0.7701 0.9088 1.8799
##
## sigma^2 estimated as 4.911: log likelihood = -18.08, aic = 44.16Sometimes, we just want to keep the coefficient.
coef(ARMA11) #Get coefficient## ar1 ma1 intercept
## 0.3551015 0.4889285 6.6417184The following code shows the plot of residuals.
plot.ts(ARMA11$residuals) 
R has a convenient function to autofit() the parameter of ARIMA model. This requires the forecast package. The following code installs and loads the forecast package:
install.packages("forecast")
library(forecast)Now we are ready to find the best ARIMA model.
autoarma <-auto.arima (price)
autoarma## Series: price
## ARIMA(0,1,0)
##
## sigma^2 estimated as 6.429: log likelihood=-16.45
## AIC=34.89 AICc=35.69 BIC=34.84One important function of time series model is forecasting. The following code gives the two-step ahead forecast:
theForecast <-predict(ARMA11,
n.ahead=2,
se.fit=TRUE) The following shows the forecast plot.
plot.ts(theForecast$pred) #visualize the forecast
GARCH
To estimate ARCH and GARCH model, we need to install and load package rugarch.
install.packages("rugarch")
library(rugarch)We will estimate GARCH(1,1) with ARMA(1,1) on generate random number
a <- runif(1000, min=0, max=100) #random number
Spec <-ugarchspec(
variance.model=list(model="sGARCH",
garchOrder=c(1,1)),
mean.model=list(armaOrder=c(1,1)),
distribution.model="std")
Garch <- ugarchfit(spec=Spec, data=a)
Garch##
## *---------------------------------*
## * GARCH Model Fit *
## *---------------------------------*
##
## Conditional Variance Dynamics
## -----------------------------------
## GARCH Model : sGARCH(1,1)
## Mean Model : ARFIMA(1,0,1)
## Distribution : std
##
## Optimal Parameters
## ------------------------------------
## Estimate Std. Error t value Pr(>|t|)
## mu 50.03765 0.870258 57.497493 0.000000
## ar1 -0.79951 0.313322 -2.551732 0.010719
## ma1 0.78029 0.326027 2.393336 0.016696
## omega 2.57816 5.152657 0.500355 0.616825
## alpha1 0.00000 0.006638 0.000012 0.999990
## beta1 0.99674 0.000413 2415.765954 0.000000
## shape 99.99959 43.561068 2.295618 0.021698
##
## Robust Standard Errors:
## Estimate Std. Error t value Pr(>|t|)
## mu 50.03765 0.879966 56.863153 0.000000
## ar1 -0.79951 0.222585 -3.591957 0.000328
## ma1 0.78029 0.234888 3.321976 0.000894
## omega 2.57816 3.462243 0.744650 0.456483
## alpha1 0.00000 0.004471 0.000018 0.999986
## beta1 0.99674 0.000151 6600.081639 0.000000
## shape 99.99959 4.129236 24.217453 0.000000
##
## LogLikelihood : -4741.296
##
## Information Criteria
## ------------------------------------
##
## Akaike 9.4966
## Bayes 9.5309
## Shibata 9.4965
## Hannan-Quinn 9.5096
##
## Weighted Ljung-Box Test on Standardized Residuals
## ------------------------------------
## statistic p-value
## Lag[1] 0.3277 0.5670
## Lag[2*(p+q)+(p+q)-1][5] 0.9018 1.0000
## Lag[4*(p+q)+(p+q)-1][9] 1.7943 0.9916
## d.o.f=2
## H0 : No serial correlation
##
## Weighted Ljung-Box Test on Standardized Squared Residuals
## ------------------------------------
## statistic p-value
## Lag[1] 0.918 0.3380
## Lag[2*(p+q)+(p+q)-1][5] 4.572 0.1908
## Lag[4*(p+q)+(p+q)-1][9] 8.297 0.1127
## d.o.f=2
##
## Weighted ARCH LM Tests
## ------------------------------------
## Statistic Shape Scale P-Value
## ARCH Lag[3] 2.146 0.500 2.000 0.14298
## ARCH Lag[5] 6.832 1.440 1.667 0.03818
## ARCH Lag[7] 7.529 2.315 1.543 0.06667
##
## Nyblom stability test
## ------------------------------------
## Joint Statistic: 180.7353
## Individual Statistics:
## mu 0.15471
## ar1 0.04098
## ma1 0.04380
## omega 0.02300
## alpha1 0.02138
## beta1 0.02306
## shape 33.46656
##
## Asymptotic Critical Values (10% 5% 1%)
## Joint Statistic: 1.69 1.9 2.35
## Individual Statistic: 0.35 0.47 0.75
##
## Sign Bias Test
## ------------------------------------
## t-value prob sig
## Sign Bias 0.7992 0.4244
## Negative Sign Bias 0.4553 0.6490
## Positive Sign Bias 1.0607 0.2891
## Joint Effect 2.1474 0.5424
##
##
## Adjusted Pearson Goodness-of-Fit Test:
## ------------------------------------
## group statistic p-value(g-1)
## 1 20 125.4 1.095e-17
## 2 30 192.7 4.342e-26
## 3 40 174.6 4.506e-19
## 4 50 229.7 3.406e-25
##
##
## Elapsed time : 0.226099To obtain specific result of the GARCH model, we use the following code:
coefficient <-coef(Garch)
volatility <- sigma(Garch)
long.run.variance <- uncvariance(Garch)VAR
The following data will be used to estimate VAR model.
a<- c(3.1,3.4,5.6,7.5,5,6,6,7.5,4.5,
6.7,9,10,8.9,7.2,7.5,6.8,9.1)
b <- c(3.2,3.3,4.6,8.5,6,6.2,5.9,7.3,
4,7.2,3,12,9,6.5,5,6,7.5)
d <-c(4,6.2,5.3,3.6,7.5,6,6.2,6.9,
8.2,4,4.2,5,11,3,2.5,3,6)
df <-data.frame(a,b,d)To estimate a VAR model, we need to install and load vars packages.
install.packages("vars")
library(vars) The following code estimates the VAR(2) model.
abvar<-VAR(df,p=2) #run VAR(2)
coef(abvar) #coefficient estiamte of VAR## $a
## Estimate Std. Error t value Pr(>|t|)
## a.l1 0.59366615 0.3776796 1.5718776 0.1546224
## b.l1 -0.53728016 0.3153665 -1.7036692 0.1268463
## d.l1 0.06427631 0.2620526 0.2452802 0.8124144
## a.l2 0.59478214 0.4406631 1.3497434 0.2140455
## b.l2 -0.49271501 0.3681374 -1.3383999 0.2175550
## d.l2 0.13247171 0.2139979 0.6190328 0.5531098
## const 4.55502873 2.5585550 1.7803130 0.1128965
##
## $b
## Estimate Std. Error t value Pr(>|t|)
## a.l1 1.03577283 0.4390665 2.3590341 0.04602757
## b.l1 -0.98047999 0.3666252 -2.6743391 0.02817211
## d.l1 0.43849071 0.3046458 1.4393458 0.18800763
## a.l2 0.77048414 0.5122871 1.5040084 0.17098968
## b.l2 -0.74120465 0.4279733 -1.7318948 0.12153171
## d.l2 -0.08577585 0.2487804 -0.3447853 0.73914487
## const 3.30736245 2.9744145 1.1119373 0.29846082
##
## $d
## Estimate Std. Error t value Pr(>|t|)
## a.l1 0.09567126 0.4508959 0.2121804 0.8372726
## b.l1 0.37908977 0.3765028 1.0068710 0.3434765
## d.l1 0.26703079 0.3128536 0.8535327 0.4181858
## a.l2 0.23097179 0.5260892 0.4390354 0.6722522
## b.l2 -0.67987321 0.4395038 -1.5469110 0.1604714
## d.l2 -0.18657598 0.2554831 -0.7302869 0.4860477
## const 4.67527066 3.0545515 1.5305915 0.1644018summary(abvar) #summary of VAR##
## VAR Estimation Results:
## =========================
## Endogenous variables: a, b, d
## Deterministic variables: const
## Sample size: 15
## Log Likelihood: -71.865
## Roots of the characteristic polynomial:
## 0.8034 0.8034 0.6583 0.6583 0.4639 0.4639
## Call:
## VAR(y = df, p = 2)
##
##
## Estimation results for equation a:
## ==================================
## a = a.l1 + b.l1 + d.l1 + a.l2 + b.l2 + d.l2 + const
##
## Estimate Std. Error t value Pr(>|t|)
## a.l1 0.59367 0.37768 1.572 0.155
## b.l1 -0.53728 0.31537 -1.704 0.127
## d.l1 0.06428 0.26205 0.245 0.812
## a.l2 0.59478 0.44066 1.350 0.214
## b.l2 -0.49272 0.36814 -1.338 0.218
## d.l2 0.13247 0.21400 0.619 0.553
## const 4.55503 2.55856 1.780 0.113
##
##
## Residual standard error: 1.552 on 8 degrees of freedom
## Multiple R-Squared: 0.4615, Adjusted R-squared: 0.0576
## F-statistic: 1.143 on 6 and 8 DF, p-value: 0.4184
##
##
## Estimation results for equation b:
## ==================================
## b = a.l1 + b.l1 + d.l1 + a.l2 + b.l2 + d.l2 + const
##
## Estimate Std. Error t value Pr(>|t|)
## a.l1 1.03577 0.43907 2.359 0.0460 *
## b.l1 -0.98048 0.36663 -2.674 0.0282 *
## d.l1 0.43849 0.30465 1.439 0.1880
## a.l2 0.77048 0.51229 1.504 0.1710
## b.l2 -0.74120 0.42797 -1.732 0.1215
## d.l2 -0.08578 0.24878 -0.345 0.7391
## const 3.30736 2.97441 1.112 0.2985
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
##
## Residual standard error: 1.805 on 8 degrees of freedom
## Multiple R-Squared: 0.616, Adjusted R-squared: 0.328
## F-statistic: 2.139 on 6 and 8 DF, p-value: 0.1578
##
##
## Estimation results for equation d:
## ==================================
## d = a.l1 + b.l1 + d.l1 + a.l2 + b.l2 + d.l2 + const
##
## Estimate Std. Error t value Pr(>|t|)
## a.l1 0.09567 0.45090 0.212 0.837
## b.l1 0.37909 0.37650 1.007 0.343
## d.l1 0.26703 0.31285 0.854 0.418
## a.l2 0.23097 0.52609 0.439 0.672
## b.l2 -0.67987 0.43950 -1.547 0.160
## d.l2 -0.18658 0.25548 -0.730 0.486
## const 4.67527 3.05455 1.531 0.164
##
##
## Residual standard error: 1.853 on 8 degrees of freedom
## Multiple R-Squared: 0.6278, Adjusted R-squared: 0.3487
## F-statistic: 2.249 on 6 and 8 DF, p-value: 0.143
##
##
##
## Covariance matrix of residuals:
## a b d
## a 2.4097 0.7713 -1.0454
## b 0.7713 3.2566 0.6701
## d -1.0454 0.6701 3.4345
##
## Correlation matrix of residuals:
## a b d
## a 1.0000 0.2753 -0.3634
## b 0.2753 1.0000 0.2004
## d -0.3634 0.2004 1.0000To generate coefficient plot, we need to install and load coefplot package:
install.packages("coefplot")
library(coefplot)The following code generates the coefficient plot for the VAR model:
coefplot(abvar$varresult$a)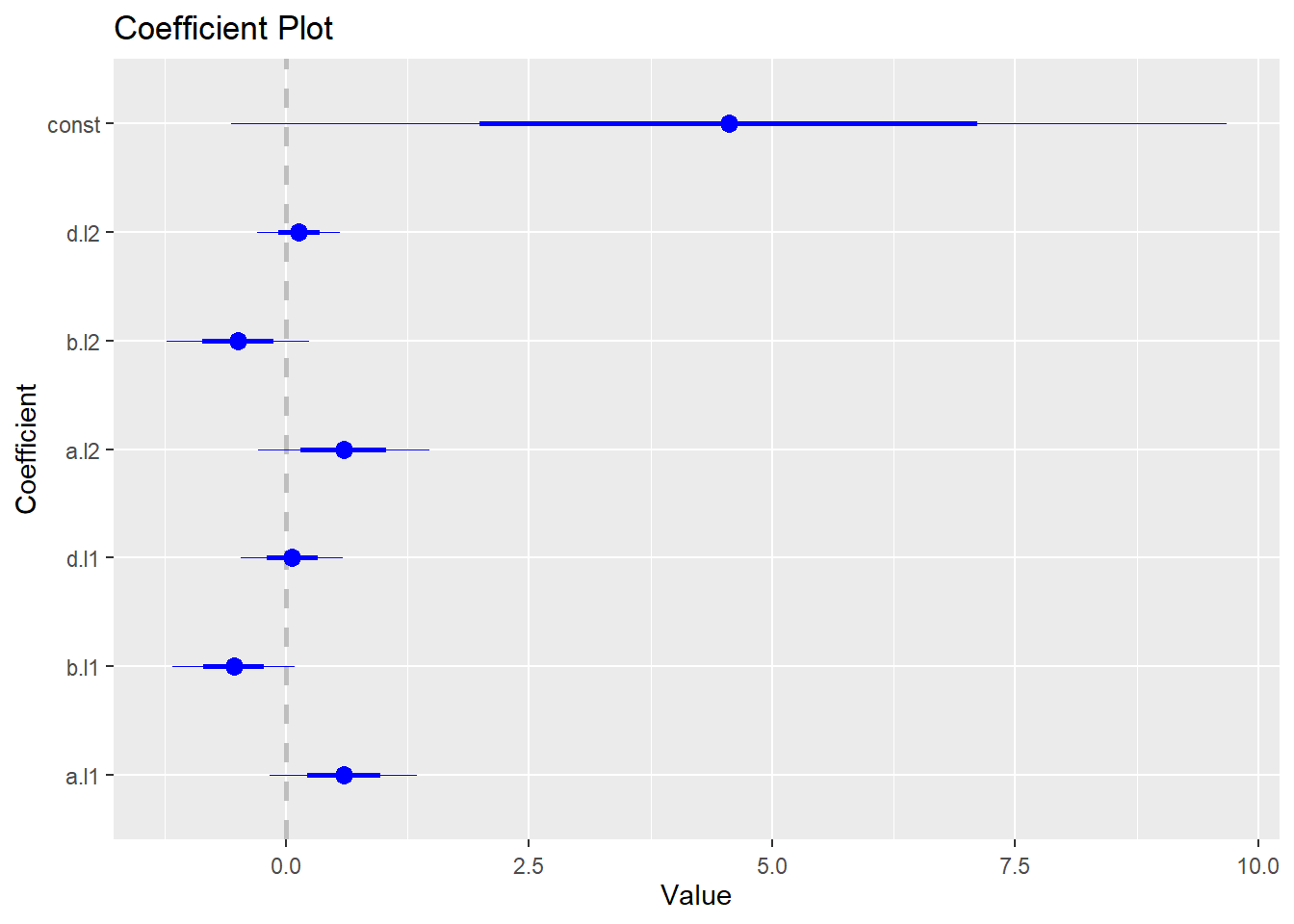
coefplot(abvar$varresult$b)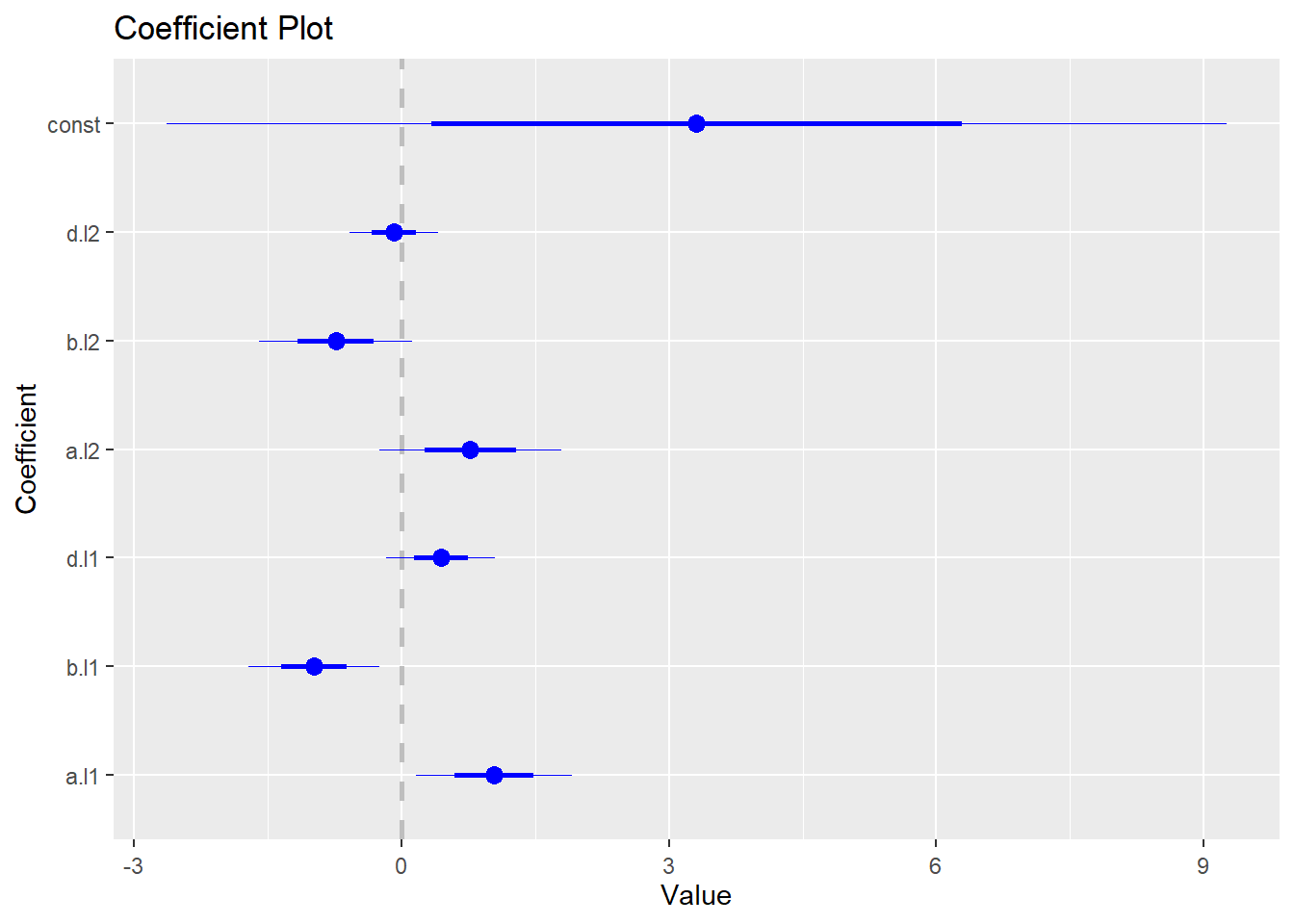
coefplot(abvar$varresult$d)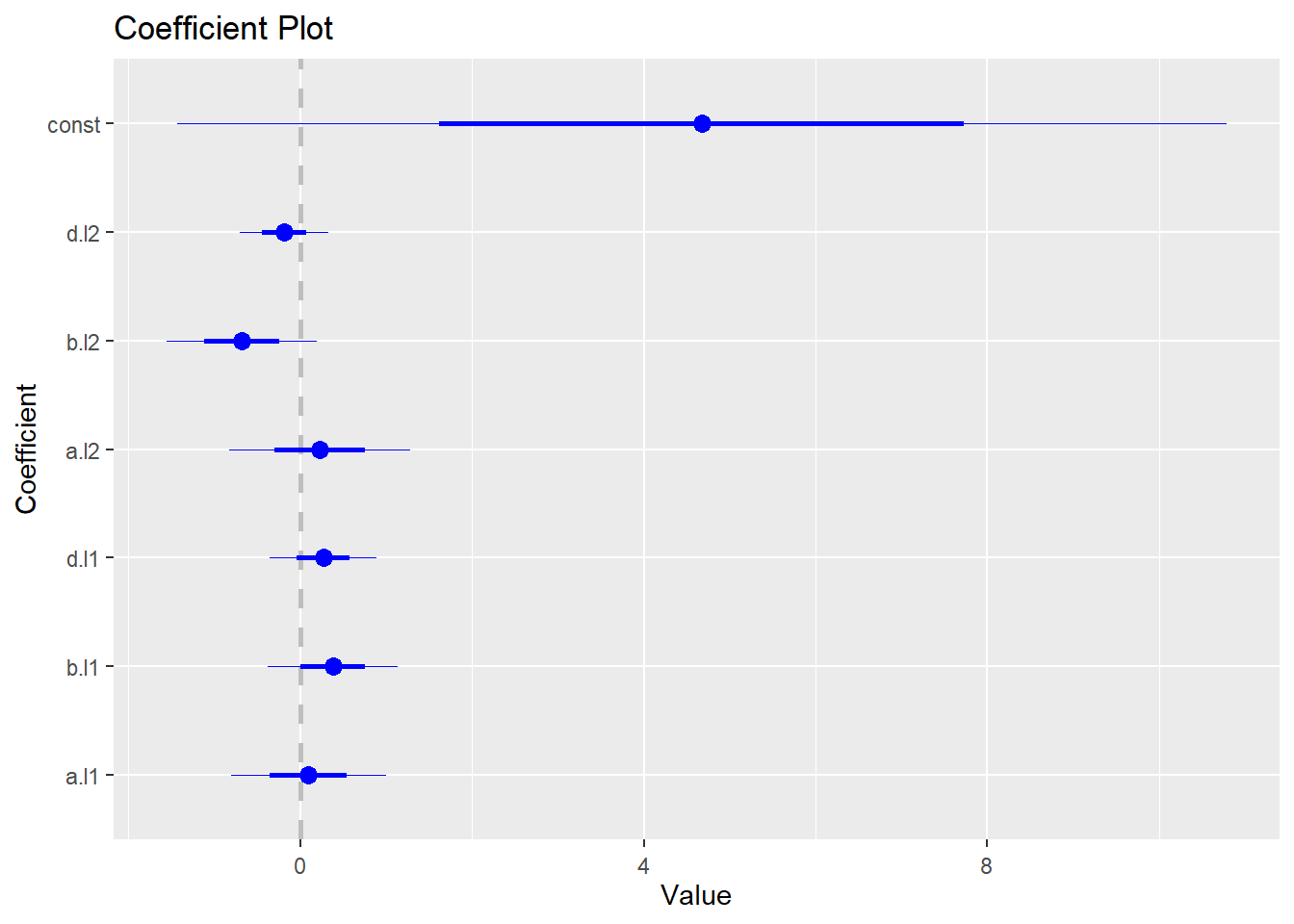
Panel Data
To estimate panel data model, we need to install and load package plm.
install.packages("plm")
library(plm)We have the following panel data
time <- c(1,2,1,2,1,2,1,2,1,2,1,2)
firm <-c(1,1,2,2,3,3,4,4,5,5,6,6)
x <- c(1, 2, 4, 3, 5, 4, 2, 5,5,6,7,8)
y <- c(2, 3, 5, 4, 3, 2, 4, 7,7,8,5,7)
p.df <- data.frame(time, firm, x, y)
p.df <- pdata.frame(p.df, index = c("firm", "time"), drop.index = F, row.names = T)Clustering
We first estimate the model based on pooled OLS.
pooled.plm <- plm(formula=y~x, data=p.df, model="pooling") Then we calculate the variance-covariance matrix to be clustered by group.
coeftest(pooled.plm, vcov=vcovHC(pooled.plm,
type="sss",
cluster="group")) ##
## t test of coefficients:
##
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.81164 0.68521 2.6439 0.024568 *
## x 0.67808 0.17151 3.9536 0.002715 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Then we calculate the variance-covariance matrix to be clustered by time.
coeftest(pooled.plm, vcov=vcovHC(pooled.plm,
type="sss",
cluster="time")) ##
## t test of coefficients:
##
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.81164 0.71388 2.5377 0.029478 *
## x 0.67808 0.21088 3.2154 0.009246 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Two-way clustering of both time and group:
coeftest(pooled.plm, vcov=vcovDC(pooled.plm,
type="sss"))##
## t test of coefficients:
##
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.81164 0.81130 2.2330 0.04959 *
## x 0.67808 0.22838 2.9691 0.01407 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Fixed Effect Model
We first estimate the model based on pooled OLS.
fe.plm <- plm(formula=y~x, data=p.df, model="within")Then we calculate the variance-covariance matrix to be clustered by group.
coeftest(fe.plm, vcov=vcovHC(fe.plm,
type="sss",
cluster="group")) ##
## t test of coefficients:
##
## Estimate Std. Error t value Pr(>|t|)
## x 1.071429 0.089074 12.028 7.008e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Then we calculate the variance-covariance matrix to be clustered by time.
coeftest(fe.plm, vcov=vcovHC(fe.plm,
type="sss",
cluster="time")) ##
## t test of coefficients:
##
## Estimate Std. Error t value Pr(>|t|)
## x 1.0714e+00 4.6089e-10 2324719242 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Simulation
We will consider two different simulations:
- random walk model, and
- Roll model of security price.
Random Walk
We first construct a random walk function that simulates random walk model. It takes the number of period (N), initial value (x0), drift (mu), and variance. The function use rnorm() to generate random normal variable, and then use cumsum() to get the random walk. Note that the whole function is based on vector operation, instead of looping over time.
RW <- function(N, x0, mu, variance) {
z<-cumsum(rnorm(n=N, mean=0,
sd=sqrt(variance)))
t<-1:N
x<-x0+t*mu+z
return(x)
}
# mu is the drift
P1<-RW(100,10,0,0.0004)
P2<-RW(100,10,0,0.0004)
plot(P1, main="Random Walk without Drift",
xlab="t",ylab="Price", ylim=c(9.7,10.3),
typ='l', col="red")
par(new=T) #to draw in the same plot
plot(P2, main="Random Walk without Drift",
xlab="t",ylab="Price", ylim=c(9.7,10.3),
typ='l', col="blue")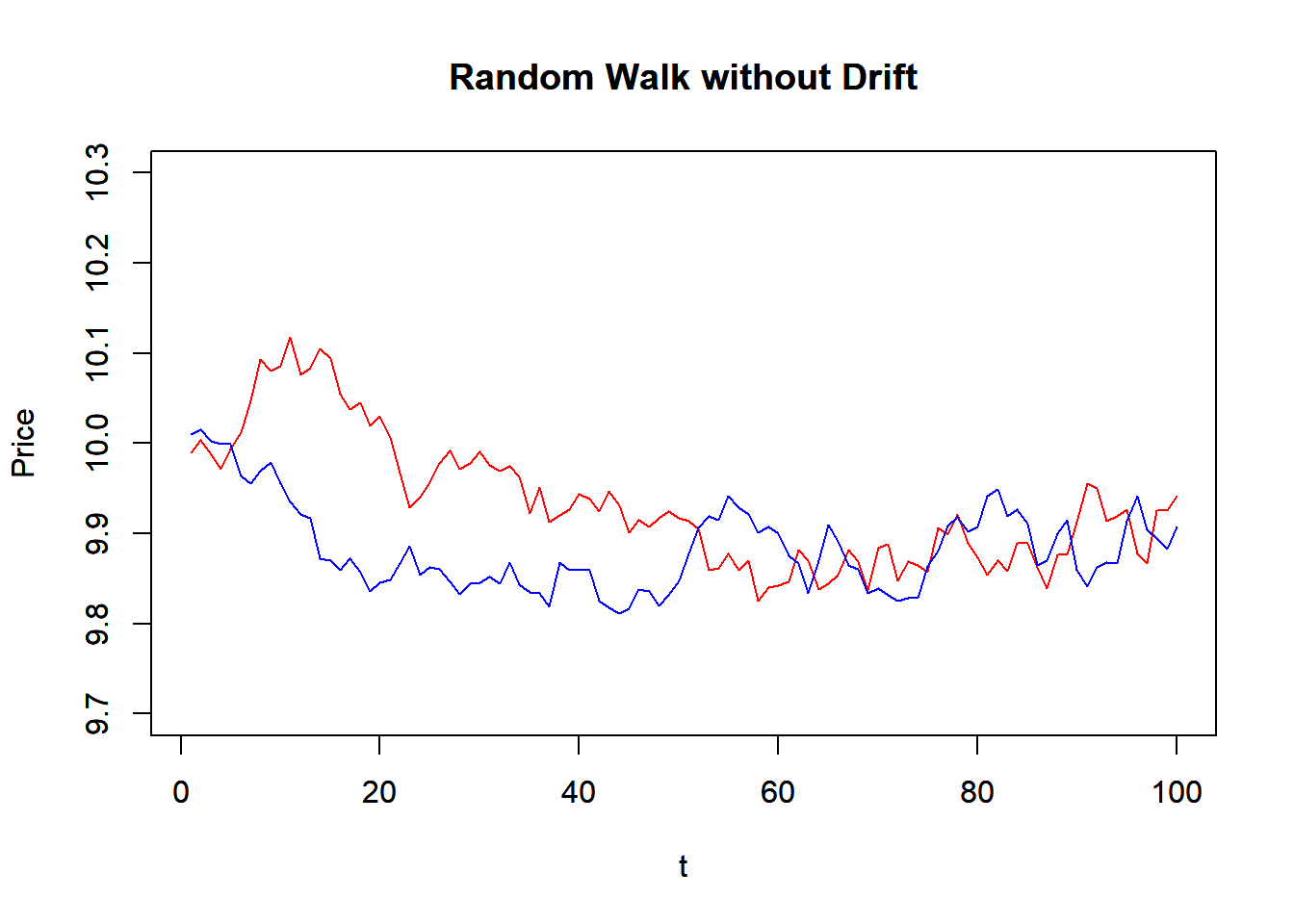
par(new=F)Roll Model
To simulate the Roll model, we first simulate the price series and simulated trade prices. We calculate the first difference of the price and then we can estimate the first and the second orders of autocovariances. Then we can compare the true trading cost with the estimate one.
require(zoo)
trial <-1000 #Number of trial
cost <-c() #cost each trial
sd <-c() #sd each trial
true.cost = 2
true.sd = 1
time = 1:trial
for (i in 1:trial) {
#Simulated Price Series
epsilon = rnorm(time,sd=true.sd)
prices = cumsum(epsilon)
m_t = zoo(prices)
a_t = m_t + true.cost
b_t = m_t - true.cost
#simulated trade prices
q_t = sign(rnorm(time))
p_t = m_t + (true.cost * q_t)
#1st difference of prices
delta_p <- p_t-lag(p_t)
#omit n.a. entry
delta_p <- na.omit(delta_p)
gamma_0 <- var(delta_p)
gamma_1 <- cov(delta_p[1:length(delta_p)-1],
delta_p[2:length(delta_p)])
sigma_2 <- gamma_0 + 2 * gamma_1
if(gamma_1 > 0){
print("Error: Positive Autocovariance!")
} else {
cost <- append(cost,sqrt(-1*gamma_1))
sd <-append(sd,sigma_2)
}
}
# Stimulated Cost Plot
plot(cost)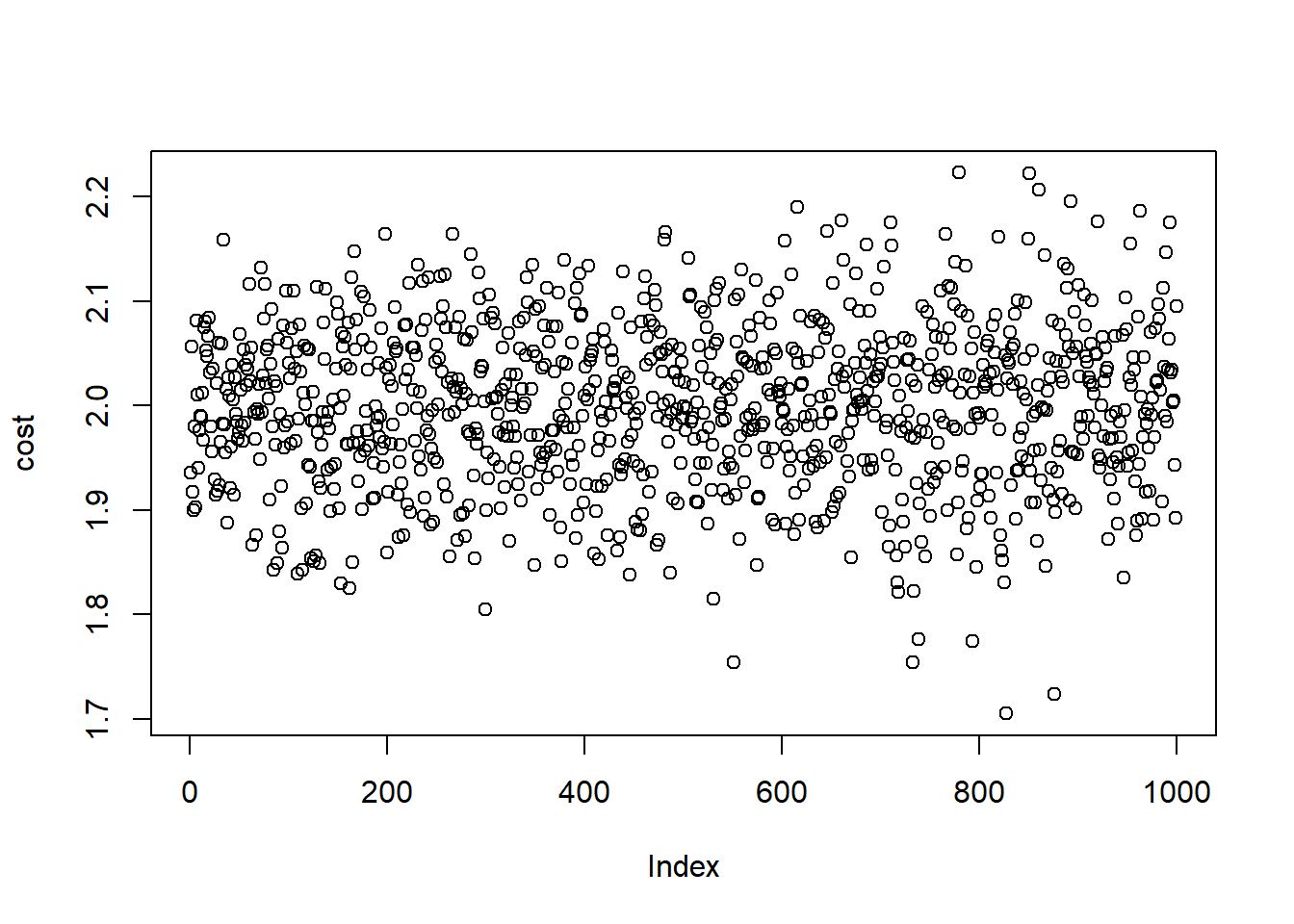
est.cost <- mean(cost)
plot(sd)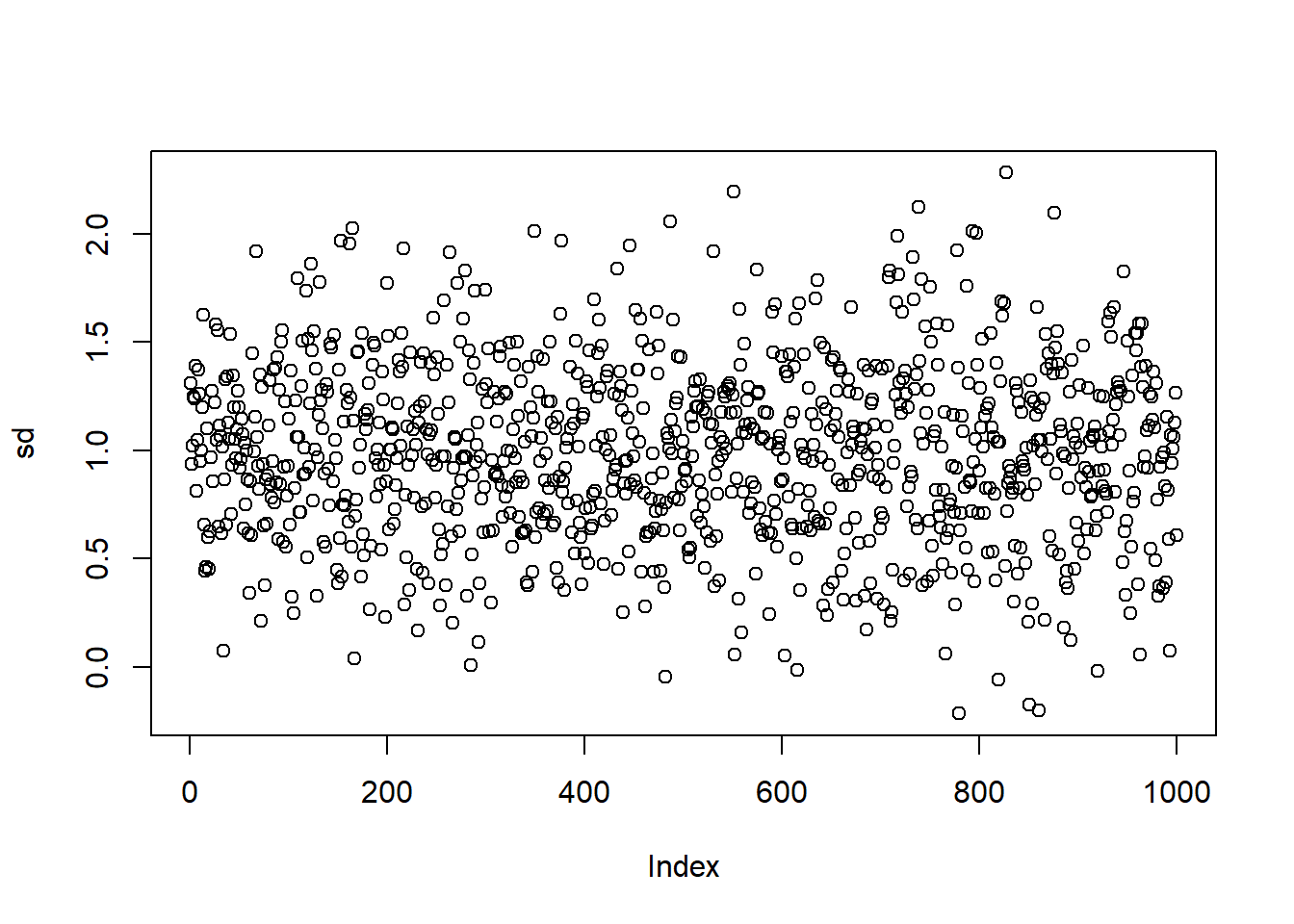
est.sd <- mean(sd)
# Final Result
cat("True cost and sd are", true.cost," and ", true.sd)## True cost and sd are 2 and 1cat("Estimated cost and sd are", est.cost," and ", est.sd)## Estimated cost and sd are 1.999111 and 0.9916815